What is artificial intelligence?
Artificial intelligence (AI) employs computer systems to perform tasks that normally require human intelligence, such as speech recognition and visual perception. AI relies on technologies and algorithms such as robotics, machine learning, and the internet to imitate the workings of the human brain. With unlimited computational power and storage capacity, AI has the potential to outperform human beings.
In medicine, computer vision algorithms have the potential to recognise abnormalities and diseases by evaluating colour, shape, and patterns [1].
Examples of AI applications include:
- Technology to enable self-driving cars
- Speech recognition algorithms to interact with humans, such as Apple's SIRI, Amazon's Alexa, and Google Assistant
- Language translation algorithms
- Identification of dog breeds (one algorithm has been reported to have achieved an accuracy of more than 96%)
- Prediction of user preferences such as a list of movies or targeted advertisements
- Prediction of periods of high demand for a taxi or a flexible workforce [2,3].
How are artificial intelligence and deep learning used in medicine?
Deep learning computer algorithms are based on convolutional neural networks. Neural networks are based on a computational model inspired by the workings of the biological brain. A large number of connected nodes called artificial neurones are similar to biological neurones in the brain. These systems learn the features of an object by evaluating manually labelled data, such as ‘dog’ or ‘no dog’. The learned features can then be used to infer the nature of a new image.
Images are widely used to diagnose injury and disease and in studies of human anatomy and physiology. Advanced medical imaging techniques include magnetic resonance imaging (MRI), dual-energy X-ray absorptiometry, ultrasonography, and computed tomography (CT) [4–10].
In medical imaging, convolutional neural networks are used to determine ‘abnormal’ or ‘normal’. They train on large labelled databases of medical images and match or exceed human vision for the detection of objects in the images in areas such as:
- Breast cancer
- Brain tumour
- Skin cancer
- Alzheimer disease [11,12].
These computer algorithms will be scalable to multiple devices, platforms, and operating systems, reducing their cost and increasing their availability for diagnosis and research. Universities, governments, and research-funding agencies have recognised the opportunities to improve early diagnosis of diseases, such as cancer, heart disease, diabetes, and dementia, and are investing heavily in the sector.
AI techniques approved by the US Food and Drug Administration (FDA) for clinical use by September 2018 include products to:
- Identify signs of diabetic retinopathy in retinal images
- Recognise signs of stroke in CT scans
- Visualise blood flow in the heart
- Detect skin cancer from clinical images captured using a mobile app.
How is artificial intelligence used in skin cancer diagnosis?
According to the US Skin Cancer Foundation, more people are diagnosed with skin cancer each year in the US than all other cancers combined [13]. Skin cancers are commonly classified as melanoma or non-melanoma skin cancer (the keratinocytic cancers, basal cell carcinoma, and squamous cell carcinoma). Skin cancers can be difficult to distinguish from common benign skin lesions, and the appearance of melanoma is especially variable. This means that:
- Skin cancers can be missed because they are thought to be harmless
- Large numbers of harmless lesions are unnecessarily surgically excised so as not to miss a potentially dangerous cancer.
Dermatologists examine skin lesions by visual inspection and dermoscopy. They use their experience in pattern recognition to determine which skin lesions should be excised for diagnosis or treatment. In recent years, there has been a huge interest in using AI algorithms to aid lesion diagnosis. There are a number of datasets of skin lesions that are publicly available to aid AI research.
Researchers at the University of Stanford performed a dermatologist-level classification of skin cancers with a deep learning algorithm on a dataset of 129,450 clinical images that included 2,032 skin diseases [14]. They also tested their algorithm against 21 board-certified dermatologists and found the algorithm’s performance at classification was on a par with their experts.
The International Skin Imaging Collaboration (ISIC) offers an extensive public dataset that in September 2018 had 23,906 digital dermoscopic images of more than 18 types of skin lesion. Since 2016, ISIC has also conducted a yearly 'Skin lesion analysis towards melanoma detection’ challenge. The 2017 winner of their challenge achieved more than 98% accuracy in distinguishing melanomas from benign moles [15]. ISIC then included more categories of skin lesions in the 2018 challenge, such as basal cell carcinoma and actinic keratosis. We can expect improved accuracy and more categories of skin lesions to be added to the competition every year.
Machine learning algorithms for skin lesions
To create a new machine learning skin cancer algorithm, each type of skin lesion is assigned a class. At its simplest, there may be just two classes; for example, ‘benign’ and ‘malignant’, or ‘naevus’ and ‘melanoma’. More sophisticated algorithms can assess multiple classes.
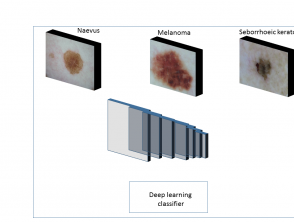
Before an algorithm is tested using a new image, deep learning algorithms are trained on a large number of images in each class. The process involves three main stages (figure 1).
Stage 1
In stage 1, the algorithm is fed with digital macroscopic or dermoscopic images labelled with the 'ground truth'. (The ground truth in this context is the lesion diagnosis, which is assigned by an expert dermatologist or is the result of histopathological examination.)
Figure 1. Overview of training of different types of skin lesions with the help of deep learning
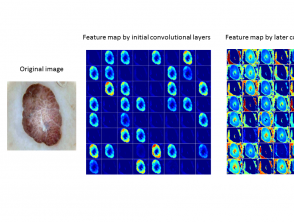
Stage 2
In stage 2, convolutional layers (a series of filters applied to the input, such as an image) extract the feature map from the images. A feature map represents data with multiple levels of abstraction.
- Initial convolutional layers extract low-level features like edges, corners, and shapes.
- Later convolutional layers extract high-level features to detect the type of skin lesion (figure 2).
Figure 2. Typical feature maps learned using convolutional neural networks
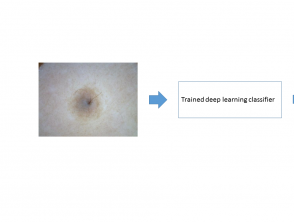
Stage 3
In stage 3, the feature maps are used by the machine learning classifier for pattern recognition of different classes of skin lesion. The deep learning algorithm can now be used to classify a new image (figure 3).
Figure 3. Inference produced by deep learning algorithms on a new image of a skin lesion
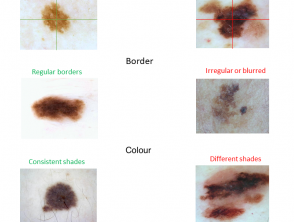
ABCD criteria
The clinical ABCD criteria used by non-experts to screen pigmented skin lesions are Asymmetry, Border irregularity, Colour variation, and Diameter over 6 mm (figure 4). See ABCDEs of melanoma, which includes 'E' for Evolution.
A: The asymmetry property checks whether two halves of the skin lesion match (or not) in terms of colour and shape. The lesions are divided into two halves based on the long axis and divided again based on the short axis. Melanoma is likely to have an asymmetrical appearance.
B: The border property defines whether the edges of skin lesion are smooth and well defined or not. Skin cancers tend to have irregular borders.
C: The colour property assesses the number and variability of colours throughout the skin lesion. Melanoma and pigmented basal cell carcinoma often include shades of 3–6 colours (black, tan, dark brown, grey, blue, red, and white), whereas naevi and freckles tend to have only one or two colours, which are symmetrically distributed.
D: The diameter property measures the approximate diameter of the skin lesion. The diameter of malignant skin lesions is generally greater than 6 mm (the size of a pencil eraser).
Figure 4. The ABCD rule for skin cancer
Yang and colleagues proposed to adopt the ABCD rule for image processing and machine learning algorithms [16]. In their work, they compared the performance of their system with doctors (general, junior, and expert) and deep learning algorithms for the diagnosis of skin lesions testing dataset (table 1). They invited two doctors from each category to perform this task.
They trained their system on their skin disease SD-198 dataset of 6,584 clinical images from 198 different lesion categories, and they extracted low-level features from three visual components: texture, colours, and borders. Yang’s computer-assisted device (CAD) system performed better than the VGG-Net and ResNet deep learning algorithm and was comparable with the performance of junior doctors. However, the expert dermatologists were significantly superior to the CAD system.
| Methods | Accuracy | Standard error | |
|---|---|---|---|
| Yang CAD system | 56.47 | 53.15 | |
| Medical experts | General doctors (n=2) | 49.00 | 47.50 |
| Junior doctors (n=2) | 52.00 | 53.40 | |
| Expert dermatologists (n=2) | 83.29 | 85.00 | |
| Deep learning | VGGNet | 50.27 | 48.25 |
| ResNet | 53.35 | 51.25 | |
Other machine learning research on skin cancer
IBM is also working on an AI tool called Watson to analyse skin lesion images for the detection of melanoma. Their device uses six key points to analyse and determine the probability of melanoma: colour, border irregularity, asymmetry level, globule and network, similarity to skin lesion images in their database, and melanoma score; these criteria are similar to ABCD criteria [17].
MetaOptima Technology Inc. has launched their DermEngine platform to provide a teledermatology service. Their visual search tool compares a user-submitted image with similar images in a database of thousands of pathology-labelled images gathered from expert dermatologists around the world. Deep learning techniques are used to search for related images based on visual features such as colour, shape, and pattern [18].
What is the future of artificial intelligence and skin cancer diagnosis?
Research involving AI is making encouraging progress in the diagnosis of skin lesions. However, AI is not going to replace medical experts in the near future. In the first place, a human is needed to select the appropriate lesion for evaluation — often among hundreds of unimportant ones.
Medical diagnosis relies on taking a careful medical history and perusal of the patient’s records. It takes into account the patient's ethnicity, skin, hair and eye colour, occupation, illness, medicines, existing sun damage, the number of melanocytic naevi, and lifestyle habits (such as sun exposure, smoking, and alcohol intake). The behaviour and previous treatment of the lesion are also clues to the diagnosis.
AI can offer a second opinion and can be used to screen out an entirely benign lesion, such as a melanocytic naevus that is symmetrical in colour and structure.
These algorithms will inevitably evolve with improved accuracy in the detection of potentially malignant skin lesions, as databases expand to include more images and more patient and lesion-specific labels.