What are chromosomes?
Chromosomes carry the genetic material of an organism (deoxyribonucleic acid [DNA]). There are 23 pairs of chromosomes in human cells. These chromosomes carry all the genetic information needed for each cell in the body to function properly.
Abnormalities in chromosomes can cause a variety of genetic disorders that can lead to developmental delay, congenital defects, and abnormal bodily function [1].
These genetic diseases can be due to either the loss or gain of chromosomes or where there are deletions or duplications of a chromosome. In these instances, a cell is missing specific genes and so cannot make certain proteins and carry out its function correctly. Examples relevant to dermatology include:
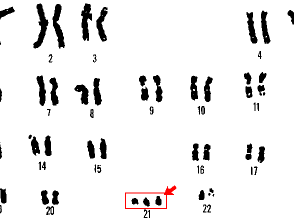
- Down syndrome (trisomy 21, where there are three copies of chromosome 21)
- Turner syndrome (a single functional X chromosome)
- DiGeorge syndrome (deletion of genes on chromosome 22).
What is cytogenetic testing?
Cytogenetics is the study of chromosomes and their structure [2].
Cytogenetic testing involves the analysis of cells in a sample of blood, tissue, amniotic fluid, bone marrow, or cerebrovascular fluid to identify any changes in an individual’s chromosomes.
There are three major methods of cytogenetic testing:
- Routine karyotyping
- Fluorescent in situ hybridisation (FISH)
- Comparative genomic hybridisation (CGH) and array comparative genomic hybridisation (aCGH).
Karyotyping
Karyotyping was one of the first methods of chromosome analysis. This method uses light microscopy and standardised staining procedures on cells in the metaphase portion of the cell cycle, when the chromosomes are lined along the equator of the cell prior to separation and are most condensed [3].
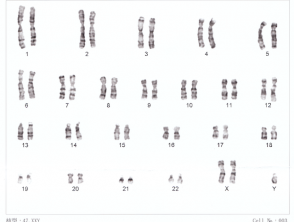
To make the chromosomal analysis more effective and efficient, stains have been developed to bind with DNA and produce characteristic banding patterns to identify different chromosomes [3]. The stain most commonly used is Giemsa dye [3]. Through this process, the chromosomes can be organised into a karyogram of 23 pairs and any abnormality involving aneuploidy (an abnormal amount of chromosomes) and large translocations (where parts of chromosomes are transplanted between each other) can be identified.
Karyotyping can only identify changes over approximately 3 megabases in size [3]. Any abnormality involving less than this will not be picked up by routine karyotyping. Karyotyping can be used to identify Down syndrome and Turner syndrome.
Karyotype for Down syndrome
Fluorescent in situ hybridisation
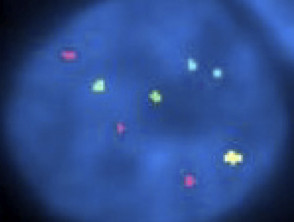
FISH was first introduced in the late 1980s and has rapidly become a well-known diagnostic cytogenetic test in both congenital and acquired disease [4]. FISH has a much higher resolution than routine karyotyping [3], especially when used on interphase cells (the phase cells remain in when not in mitosis) [5].
FISH uses fluorescent probes with complementary base sequences to locate the presence or absence of specific portions of DNA on chromosomes [2,6]. The probe and target DNA must be denatured with heat or chemicals to break hydrogen bonds in the DNA and to allow hybridisation to occur once the two samples are mixed [5]. The fluorescent probes form new hydrogen bonds with their complementary base pairs on the DNA, and these can then be detected via microscopy [5].
FISH is commonly used to detect specific chromosomal deletions or translocations associated with paediatric conditions or cancers [2]. Examples include the deletion on chromosome 22 in DiGeorge syndrome and the translocation of part of a gene on chromosomes 22 and 9 in chronic myeloid leukaemia [2].
FISH is also used for melanocytic lesions to distinguish atypical melanocytic naevi (eg, Spitz naevus) from melanoma.
Fluorescent in situ hybridisation in childhood melanoma
Comparative genomic hybridisation
CGH is a method of molecular cytogenetic testing that detects chromosomal copy number variants (portions of the genome where sections of genes are doubled or tripled) without the need for cell culturing [7]. It was first developed to identify such changes in tumours [7,8].
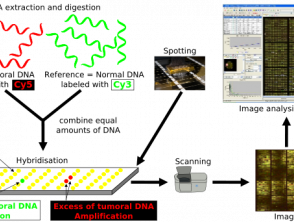
CGH uses 2 genomes; the test sample and a control, both of which are fluorescently labelled to differentiate between the two [8]. The two samples are denatured and mixed together, allowing hybridisation of metaphase chromosomes. The intensity of the fluorescent signal of the labelled test DNA relative to the control DNA can then be plotted along each chromosome, which shows the loss or gain of genetic material and allows identification of any copy number variants [8].
CGH differs from other methods of cytogenetic testing in that it does not rely on a specific target, nor does it require previous knowledge of the region under examination [8]. Instead, CGH can quickly scan a whole genome for chromosomal imbalances and it is useful in cases where the diagnosis is not known [8]. One limitation of CGH is the size of the genetic alteration that it can identify; the resolution of CGH is poor at approximately 5–10 megabases [8].
Comparative genomic hybridisation
Array comparative genomic hybridisation
aCGH utilises a similar technique as CGH but it provides a much higher resolution by using microarrays [8]. Small sections of DNA are used as targets for analysis; these sections are immobilised on solid support, which anchors DNA to a spot without altering the protein [8]. As in CGH, the sample DNA and control are fluorescently labelled to differentiate them.
The samples are then mixed and added to the microarray where they compete to bind to the probes on the microarray [8]. The strength of the different fluorescent signals can be assessed and any small gains or losses within the DNA are identified. A disadvantage of aCGH is that it cannot detect balanced chromosomal structural changes such as balanced translocations or inversions [2].
When is cytogenetic testing indicated?
Cytogenetic testing is used where a genetic abnormality is suspected.
Prenatal testing in a high risk pregnancy
Cytogenetic testing is performed in pregnancy on samples obtained in utero via amniocentesis or by chorionic villus sampling to identify a fetus with chromosomal abnormalities, such as trisomy 21 in Down syndrome.
Diagnostic testing
Cytogenetic testing is often used in paediatrics in an attempt to identify the underlying cause for developmental disorders or congenital defects. A diagnosis can be a huge relief to the families of affected children and will allow counselling regarding appropriate management and prognosis.
Haematological cancer
Cytogenetic testing is used in haematological cancers such as chronic myeloid leukaemia where a specific reciprocal translocation between chromosomes 22 and 9 results in the Philadelphia chromosome, that is present in 95% of cases.
What are the contraindications with cytogenetic testing?
There are no medical contraindications to cytogenetic testing. Informed consent should be obtained prior to arranging the tests.
What are the benefits of cytogenetic testing?
Cytogenetic testing can offer diagnosis and help with the long-term management of relevant diseases. It also allows genetic counselling for the patient or their parents about the related risk in any future pregnancies and, in some cases, guides the geneticist as to whether to test other family members.
What are the disadvantages of cytogenetic testing?
Unfortunately, cytogenetic testing is limited by its resolution. The different methods can identify small gains and losses of genetic material, as well as larger translocations, but they do not allow testing for single nucleotide variations that could contribute to the patient’s condition. There is also the possibility that cytogenetic testing will identify other chromosomal changes that are not necessarily related to the patient’s condition.